JSON file generated by CRG:
{
"ensembl_id" : "ENSG00000000003.10"
"expression_values" : [
{
"rep2_tpm" : 1.33,
"rep1_fpkm" : 6.86,
"rep1_tpm" : 2.78,
"dataset" : "ENCSR000AAA",
"rep2_fpkm" : 6.02
},
{
"rep2_tpm" : 2.09,
"rep1_fpkm" : 32.02,
"rep1_tpm" : 13.7,
"dataset" : "ENCSR000AAB",
"rep2_fpkm" : 14.48
},
...
Extensive metadata associated with each dataset (aka experiment).
Example (ENSG00000000003.10.expression.tsv):
| accession | biosample_term_name | biosample_type | developmental_slims | fpkm * | library.nucleic_acid_term_name | organ_slims | tpm * | ... |
|---|---|---|---|---|---|---|---|---|
| ENCSR000AAA | aortic smooth muscle cell | primary cell | mesoderm | 6.44 | RNA | blood vessel | 2.055 | ... |
| ENCSR000AAB | bladder microvascular endothelial cell | primary cell | mesoderm | 23.25 | RNA | blood vessel,urinary bladder | 7.895 | ... |
| ENCSR000AAC | smooth muscle cell of bladder | primary cell | mesoderm | 10.675 | RNA | urinary bladder | 2.57 | ... |
| ENCSR000AAD | bronchial epithelial cell | primary cell | endoderm | 6.355 | RNA | bronchus | 1.04 | ... |
| ENCSR000AAE | bronchial smooth muscle cell | primary cell | endoderm,mesoderm | 12.72 | RNA | bronchus | 2.995 | ... |
| ENCSR000AAF | endothelial cell of coronary artery | primary cell | mesoderm | 18.115 | RNA | blood vessel,heart | 4.965 | ... |
| ENCSR000AAG | smooth muscle cell of the coronary artery | primary cell | mesoderm | 10.775 | RNA | blood vessel,heart | 3.11 | ... |
| ENCSR000AAH | regular cardiac myocyte | primary cell | mesoderm | 16.65 | RNA | NA | 1.88 | ... |
| ENCSR000AAI | dermis blood vessel endothelial cell | primary cell | ectoderm,mesoderm | 28.08 | RNA | blood vessel,skin of body | 8.415 | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
* averaged across replicates
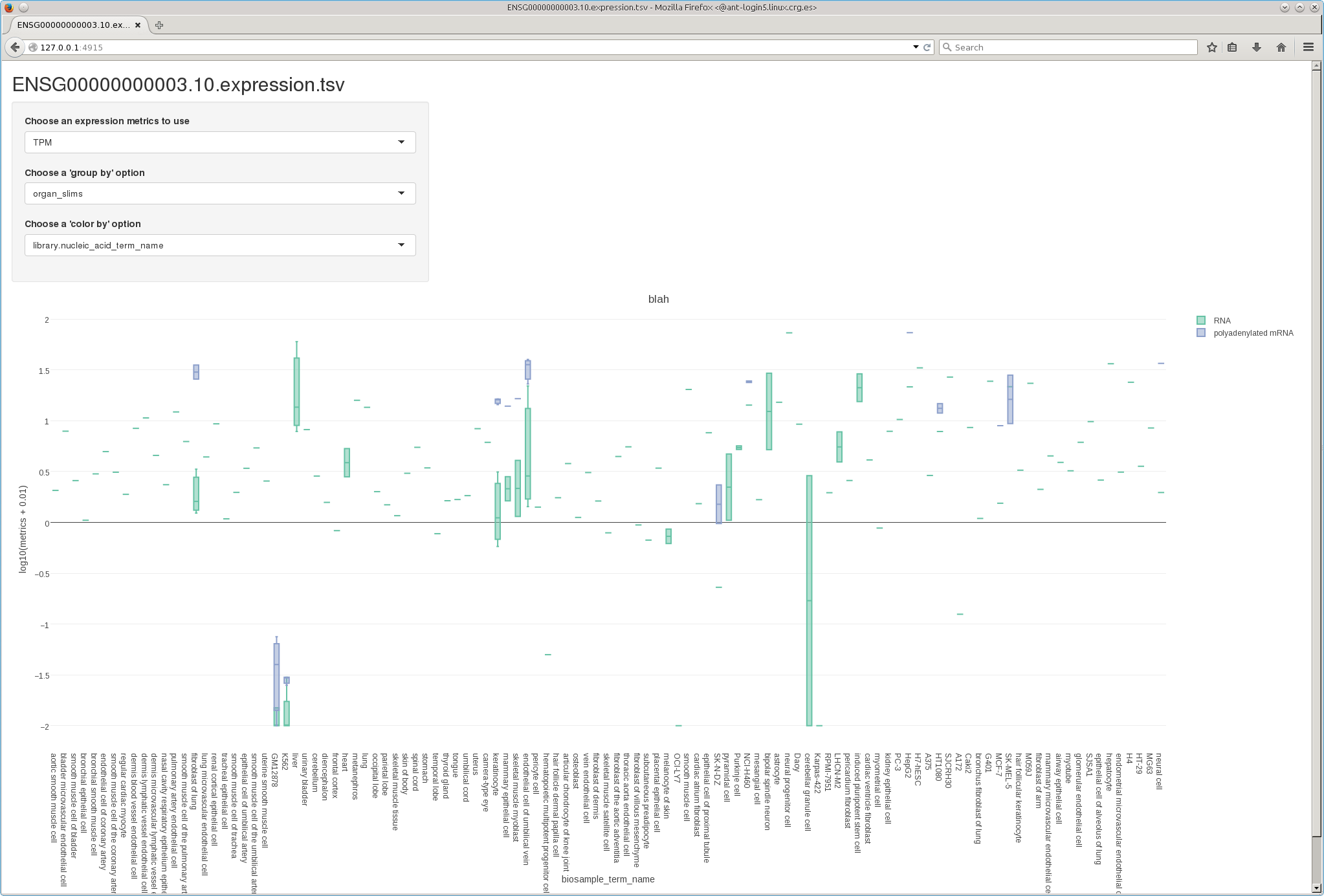
Interactive box-plots are generated by feeding the expression TSV to R's plotly library. The R script takes the following parameters:
| Parameter | Possible values |
| $geneId | Any ensembl_id |
| $metrics |
'tpm' or 'fpkm' |
| $colorBy |
Any column name of the input TSV |
| $groupBy |
Any column name of the input TSV |
Generic R code:
library(plotly) plot <- read.table("$geneId.expression.tsv", header=T, as.is=T, sep="\t") figure<-plot_ly(plot, x=$groupBy, y=log10($metrics+0.01), color=$colorBy, type="box", boxpoints = "all", jitter = 0.3, pointpos = -1.8) %>% layout( title="$geneId / $groupBy vs $colorBy", margin= list(b=300) ) htmlwidgets::saveWidget(plotly:::toWidget(figure), "./$groupBy.vs.$colorBy.$geneId.html", selfcontained = FALSE)
| Parameter | Assigned value |
| $geneId | ENSG00000000003.10 |
| $metrics |
tpm |
| $colorBy |
library.nucleic_acid_term_name |
| $groupBy |
organ_slims |
| Parameter | Assigned value |
| $geneId | ENSG00000000003.10 |
| $metrics |
tpm |
| $colorBy |
system_slims |
| $groupBy |
biosample_term_name |
| Parameter | Assigned value |
| $geneId | ENSG00000000003.10 |
| $metrics |
tpm |
| $colorBy |
organ_slims |
| $groupBy |
system_slims |
| Parameter | Assigned value |
| $geneId | ENSG00000000003.10 |
| $metrics |
tpm |
| $colorBy |
biosample_type |
| $groupBy |
organ_slims |
Plotly interactive plots embed only some data (i.e.,keeps data only for the concerned columns, and ditches everything else), so, a priori, no.
(there might be a workaround I'm not aware of, though).
I have made some tests with Shiny, another R library compatible with plotly (see screenshot below).
It seems powerful enough, unfortunately sharing options are not great.
